Nanopore signal processing
Raw signal conversion |
|---|
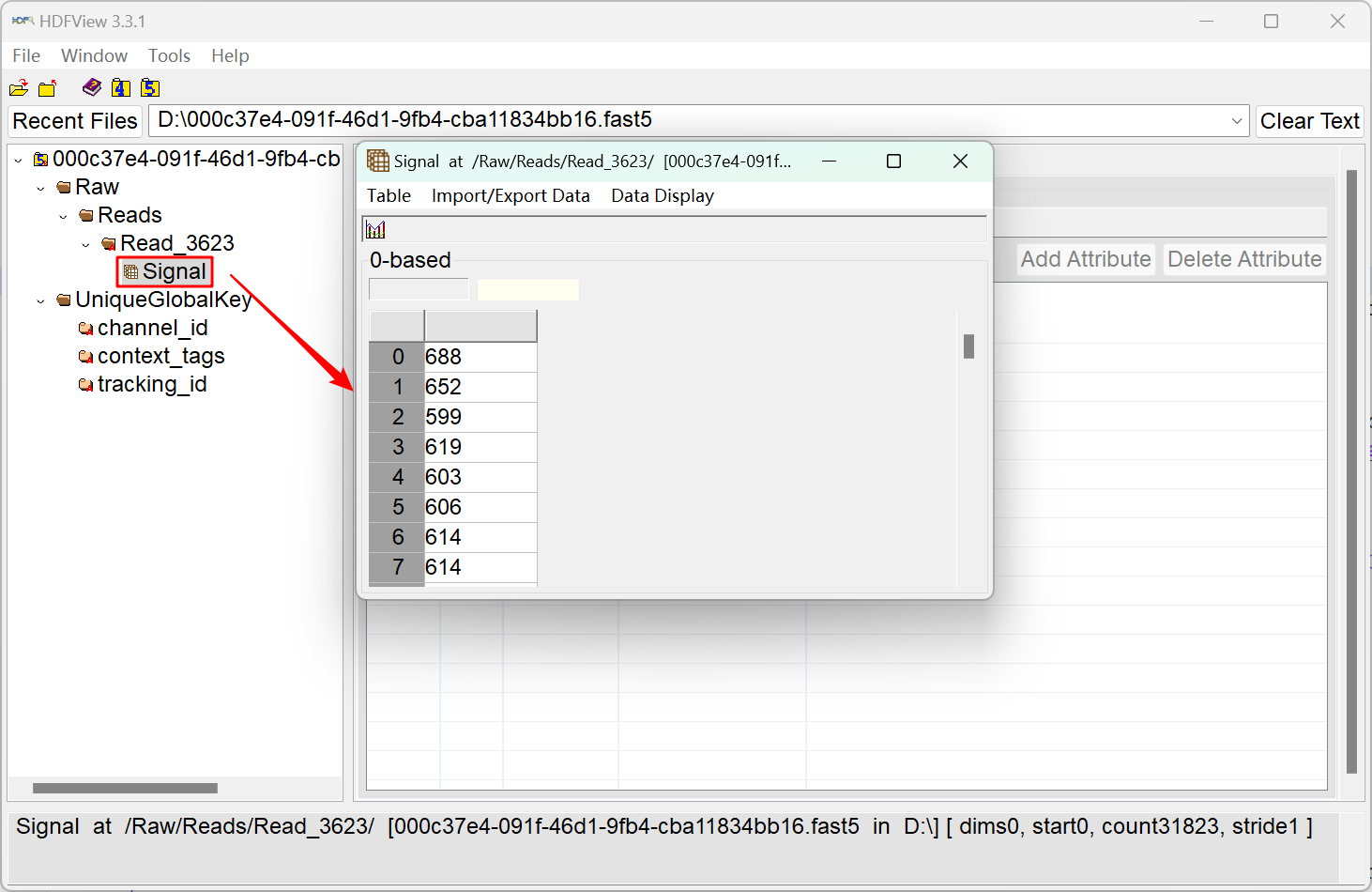
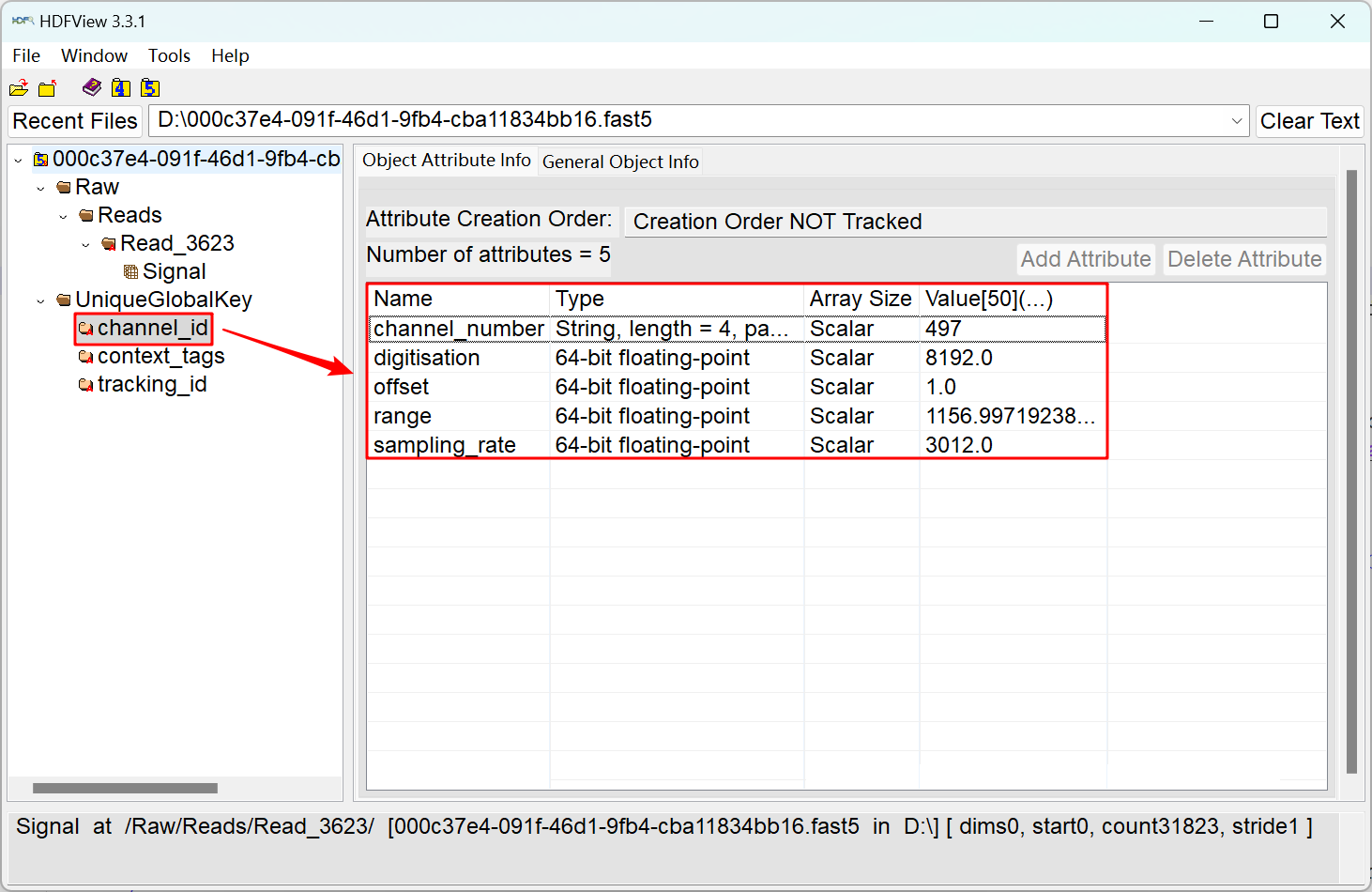
The illustration above depicts the structure of a single-fast5 file from Nanopore sequencing. In the left figure, the "Signal" data represents the current passing through the pore (type: int16). Oxford Nanopore Technology employs different pores (proteins) in various products. A single flow cell in the sequencing device can contain between 512 and 2675 pores, each referred to as a channel. As shown in the right figure, the fast5 file also includes attributes associated with the channel through which the read passes. These parameters include channel_number (the channel number from which the read was acquired), digitisation (the digitisation is the number of quantisation levels in the Analog to Digital Converter (ADC)), offset (the ADC offset error), range (the full scale measurement range in pico amperes), and sampling_rate (sampling frequency of the ADC).
The raw signal can be converted into pico Ampere (pA) values using attributes available in the channel_id group by the equation :
signal_in_pico_ampere = (raw_signal_value + offset) * range / digitisation
Signal standardization or normalization |
|---|
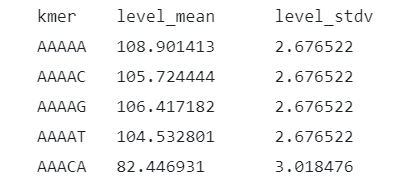
Oxford Nanopore Technologies provides "standard" parameters for each kmer [3], as illustrated in the figure below. To enhance the accuracy of analyses, some tools attempt to further standardize or normalize the signal based on these parameters. For example, Tombo employs median shift and median absolute deviation (MAD) to normalize the current signal to a 0-1 range. Nanopolish uses scaling parameters to account for per-read variations, while SegPore utilizes the polyA tail to standardize the signal for each read.
Tombo
Tombo uses median shift and MAD (median absolute deviation) scale parameters to normalize the signal:norm_signal = (signal - median) / MAD
Nanopolish
For each read, Nanopolish estimates a scale parameter to standardize the signal. You can add the --scale-events option in the nanopolish eventalign command to enable this feature. More details can be found in [6].
SegPore
SegPore first detects the polyA tail and calculates its mean (polyA_mu) and standard deviation (polyA_sigma). These values are then used to standardize the signal:stand_signal = [(signal - polyA_mu) / ployA_sigma ] * POLYA_STANDARD_SIGMA + POLYA_STANDARD_MU
Reference
[1] https://github.com/nanoporetech/taiyaki/blob/master/docs/FILE_FORMATS.md
[2] Gamaarachchi, H., Samarakoon, H., Jenner, S.P. et al. Fast nanopore sequencing data analysis with SLOW5. Nat Biotechnol 40, 1026–1029 (2022).
[3] https://github.com/nanoporetech/kmer_models
[4] https://nanoporetech.github.io/tombo/resquiggle.html
[5] https://static-content.springer.com/esm/art%3A10.1038%2Fs41587-021-01147-4/MediaObjects/41587_2021_1147_MOESM1_ESM.pdf
[6] https://static-content.springer.com/esm/art%3A10.1038%2Fnmeth.4184/MediaObjects/41592_2017_BFnmeth4184_MOESM258_ESM.pdf