Modification detection benchmark
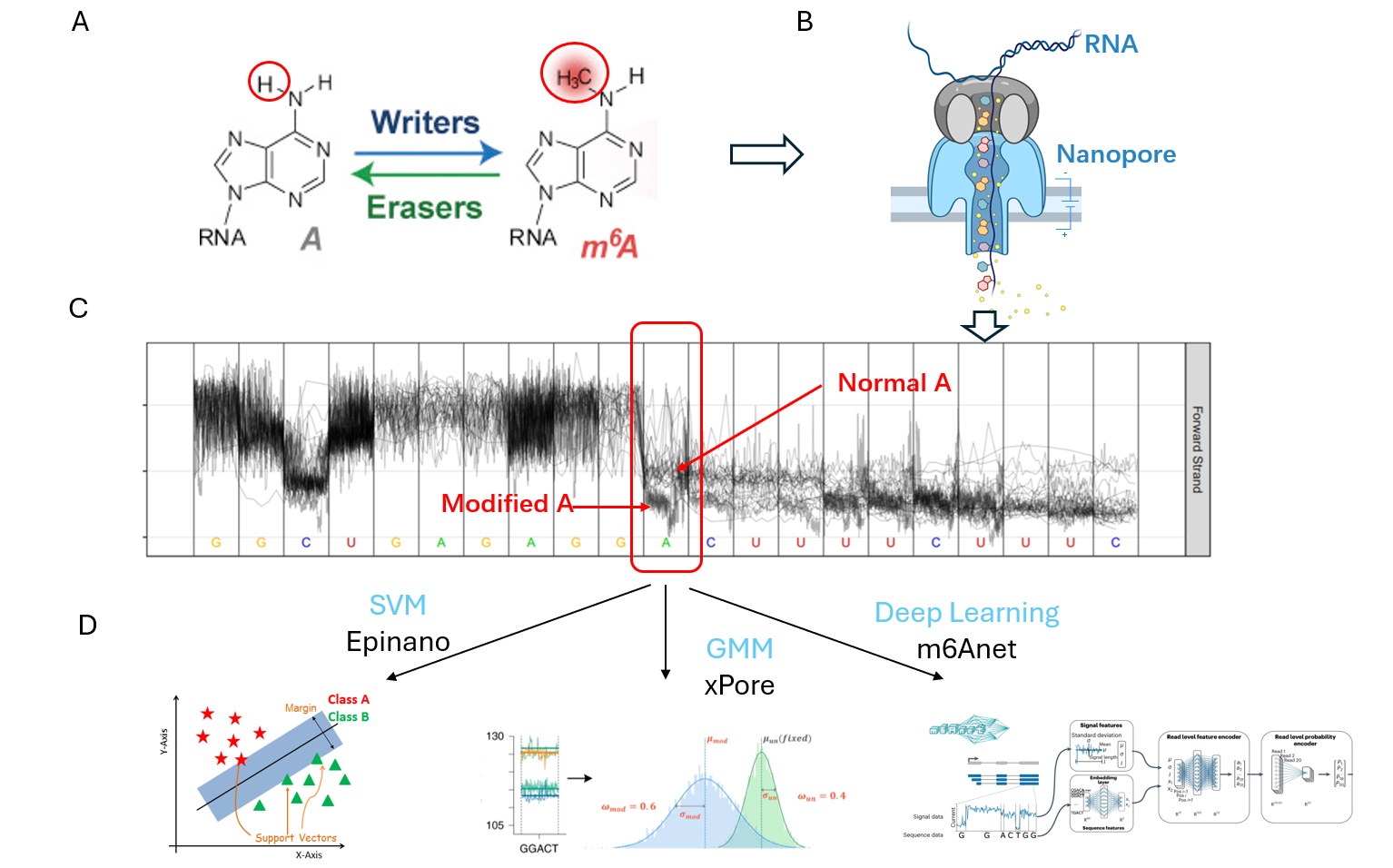
Figure A illustrates the difference between normal adenine (A) and N6-methyladenosine (m6A). After performing Nanopore sequencing, as depicted in Figure B, and subsequent analysis, the current signal variations between normal A and modified A become evident (Figure C). By analyzing these signal segments, various machine learning (ML) and deep learning (DL) models can be employed to detect the modifications.
The m6A and m5C modification detection benchmarks are available from Github.
Modification detection benchmark models introduction |
|---|
| Model | Version | Download Link | Model methods & Architecture | Limiation |
|---|---|---|---|---|
| m6Anet | 1.0 | https://github.com/GoekeLab/m6anet | MLP. | Only for m6A. |
| CHEUI | - | https://github.com/comprna/CHEUI | CNN. | Only for m6A and m5C. |
| Epinano | 1.2.0 | https://github.com/novoalab/EpiNano | SVM. | - |
| MINES | - | https://github.com/YeoLab/MINES | Random Forest | Only for m6A. |
| Nanom6A | 2.0 | https://github.com/gaoyubang/nanom6A | XGBoost. | Only for m6A. |
| SegPore | 1.0 | https://github.com/guangzhaocs/SegPore | GMM. | - |
| Tombo | 1.5.1 | https://nanoporetech.github.io/tombo | - | - |
Reference
Shi, Hailing, Jiangbo Wei, and Chuan He. "Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers." Molecular cell 74.4 (2019): 640-650.
Liu, Huanle, et al. "Accurate detection of m6A RNA modifications in native RNA sequences." Nature communications 10.1 (2019): 4079.
Pratanwanich, Ploy N., et al. "Identification of differential RNA modifications from nanopore direct RNA sequencing with xPore." Nature biotechnology 39.11 (2021): 1394-1402.
Hendra, Christopher, et al. "Detection of m6A from direct RNA sequencing using a multiple instance learning framework." Nature methods 19.12 (2022): 1590-1598.