A Multi-task Benchmark Dataset for Nanopore Sequencing
NanoBaseLib is a comprehensive dataset integrating 16 public datasets with over 30 million reads. |
||
|---|---|---|
NanoBaseLib is a benchmark platform covering 4 critical tasks. |
||
NanoBaseLib is a software package designed to incorporate new datasets efficiently. |
Principle of Nanopore sequencing
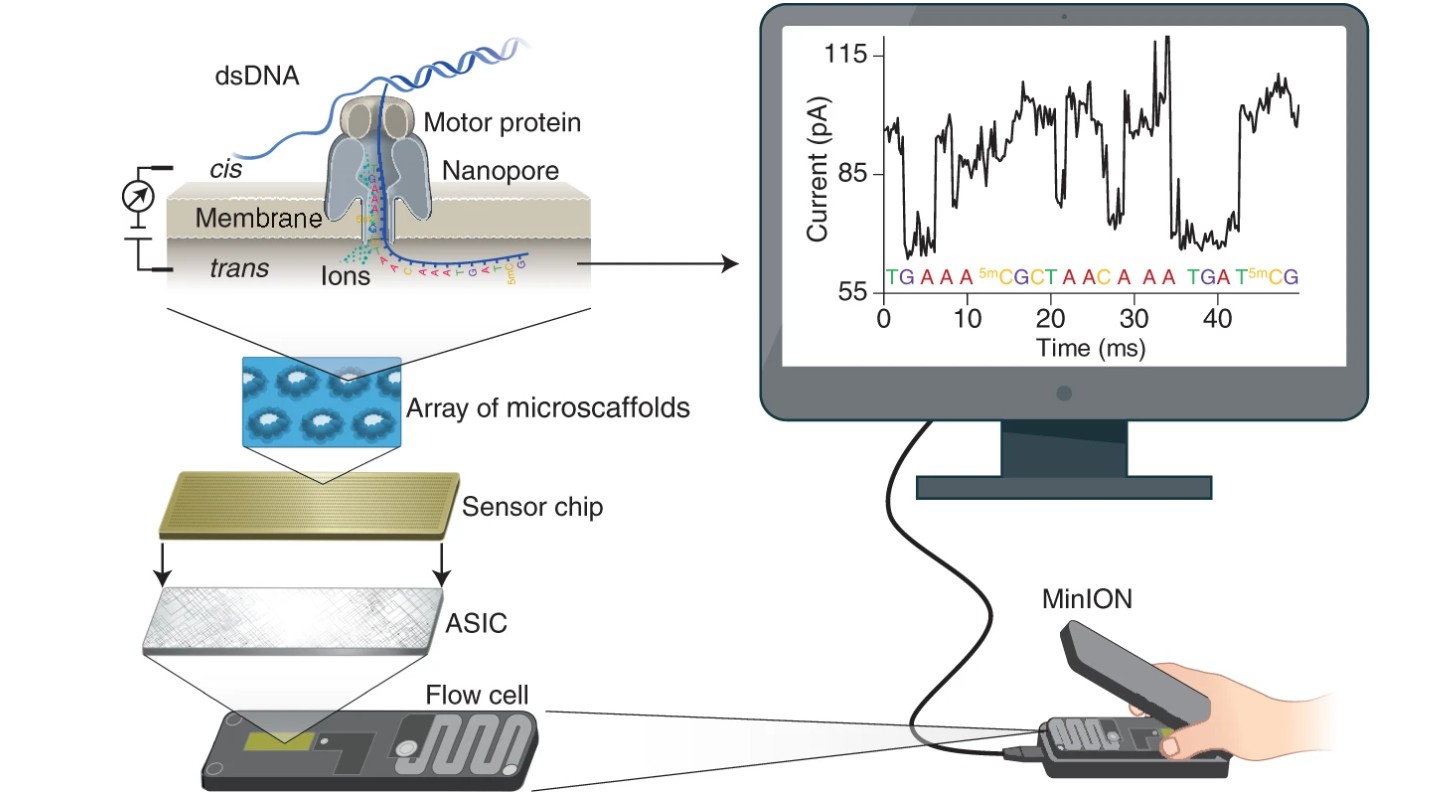
Nanopore sequencing technology measures the current signal by translocating a DNA/RNA molecule through a nanoscale pore anchored on a lipid membrane. The shape, size, and chemical properties of nucleotides in the pore jointly determine the current signals, which are collected and utilized to infer the nucleotide sequence using computational methods (e.g., seq2seq deep learning models). There are multiple nucleotides (usually 5) in the pore, which we term as kmer (e.g., 5mer). The inferred kmers are assembled into a DNA/RNA sequence computationally, which completes the sequencing process.
Beyond sequencing: machine learning powers biological discovery
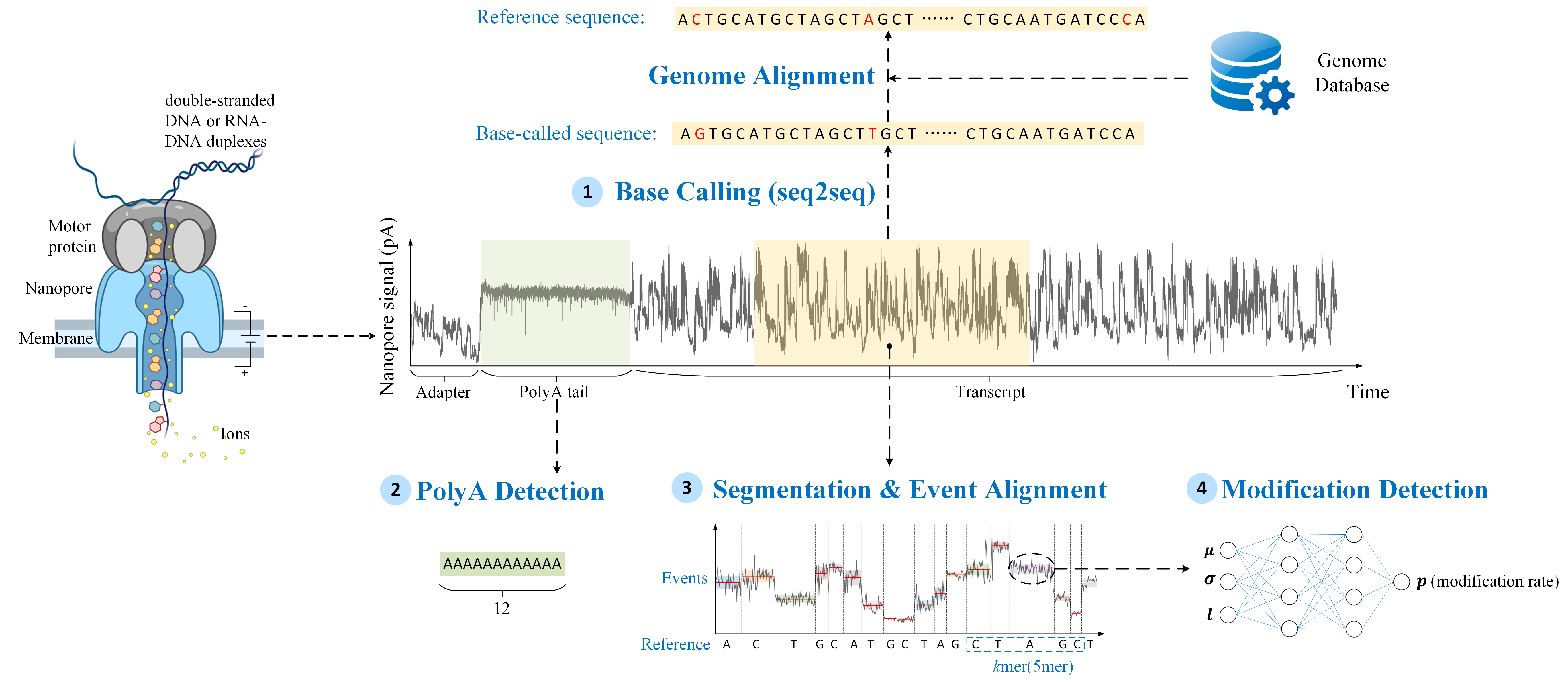
NanoBaseLib consists of four benchmark tasks: (1) Base Calling (BC), (2) PolyA Detection (PD), (3) Segmentation and Event Alignment (SA), and (4) Modification Detection (MD).
Reference
[1] Wang, Yunhao, et al. "Nanopore sequencing technology, bioinformatics and applications." Nature biotechnology 39.11 (2021): 1348-1365.